
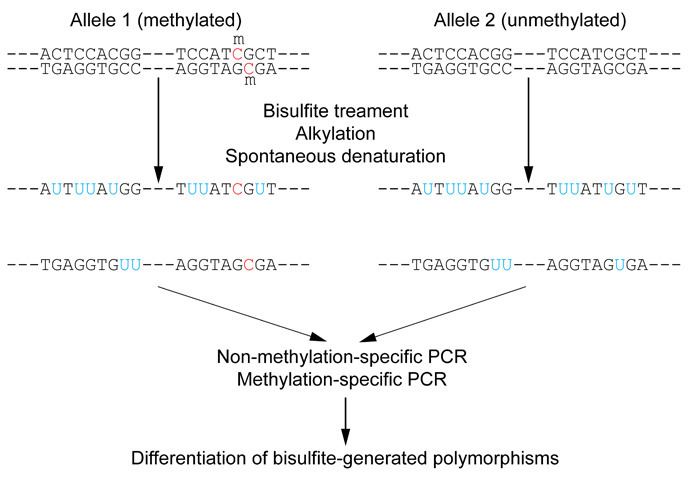

Specific enzymes, DNA methyltransferases (DNMT1, DNMT3A and DNMT3B), transfer a methyl group (CH 3) from S-adenosyl methionine (SAM) on the C5 position of the pyrimidine ring, converting cytosine (C) into 5-methylcytosine (5mC).Īmong other methods, the Bisulfite Sequencing PCR (BSP) is the most accessible and conventional method to evaluate methylation levels at single CpG resolution in a mix of DNA molecules ( Clark et al., 1994 Frommer et al., 1992). The effect of these modifications on gene transcription has been observed when several grouped CpG within a DNA region, so-called CpG islands, are modified altogether ( Greenberg and Bourc'his, 2019 Jones, 2012). DNA methylation can affect cytosine and adenine but mostly occurs on a cytosine followed by a guanine (CpG site). In vertebrates, epigenetic regulation is essential to regulate genomic imprinting, X chromosome inactivation, development regulation, cell differentiation and genome integrity preservation. Aside from transcription factor regulations, gene expression can also be activated or repressed by epigenetic modifications directly on nucleotides (DNA methylation) or histones (methylation, acetylation…).


 0 kommentar(er)
0 kommentar(er)
